Self-assembling structures of artificial proteins or how to build a molecular “origami”
If it is possible to build nanostructures of controlled shape by exploiting the double helix structure of DNA molecules (leading to assemblies commonly called “DNA origami"), it remains much more difficult to develop such structures. precise from proteins. However, in living cells, very sophisticated supramolecular architectures such as microtubules, actin filaments, or flagella perform vital functions and are entirely made up of natural proteins. The latter assemble spontaneously because each protein has a particular shape that allows it to interact in a very specific way with other proteins, thus leading to complex architectures. Creating ordered supramolecular architectures of proteins would open the door to many applications in biology but also in materials science.
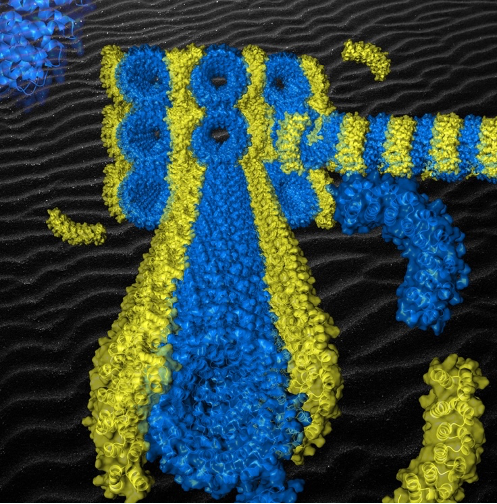
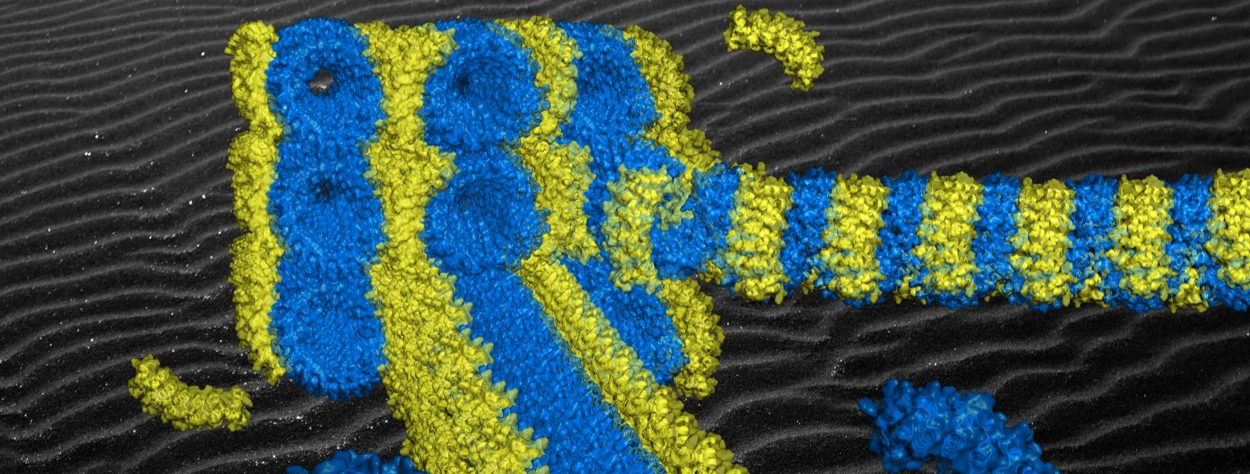
Figure: Semi-experimental model of the origami of artificial proteins whose superhelix of bricks (in blue) self-assembles by affinity with the staple (in yellow). The extreme regularity of the supramolecular structure obtained arranges the staples periodically along the superhelix and induces the formation of aligned origami crystals (see background).
@ I2BC, CEMES, CBI, IPR and ICB
- Read the article of the INP
- Design, synthesis, and characterization of protein origami based on self-assembly of a brick and staple artificial protein pair. L. Moreaud et al., PNAS, published on March 09, 2023
DOI : 10.1073/pnas.2218428120
Four French teams demonstrate the possibility of building ordered supramolecular architectures that form spontaneously from proteins specially designed for this purpose. The key to this innovation is the engineering of highly regular proteins with repeating patterns, which have recognition surfaces allowing them to establish specific interactions.
The Laboratoire Interdisciplinaire Carnot de Bourgogne (ICB), the Centre d’élaboration de matériaux et d’études structurales (CEMES, CNRS), Institut de biologie intégrative de la cellule ( I2BC, CNRS / CEA / Université Paris Saclay), l’Institut de physique de Rennes (IPR, CNRS / Université de Rennes), and the Centre de biologie intégrative (CNRS / Université de Toulouse – Paul Sabatier) have developed a generalizable method for constructing artificial protein architectures. Rather than modifying natural proteins, scientists have favored the design of new proteins that are both very regular and programmed to assemble in precise geometries capable of forming stable superstructures. One of these proteins, called “staple", has the role of arranging precisely several other proteins called “bricks", their assembly naturally giving the three-dimensional supramolecular architecture its structural complexity. A first challenge was therefore to design these proteins and produce them. Then, by combining several techniques including X-ray scattering and electron cryomicroscopy, the researchers showed that the proteins assemble in a few minutes, at room temperature and according to the expected architecture. This constitutes a first experimental whose principle can be generalized to other molecular systems.
These results are published in the journal Proceedings of the National Academy of Science.
- kc_data:
- a:8:{i:0;s:0:"";s:4:"mode";s:2:"kc";s:3:"css";s:0:"";s:9:"max_width";s:0:"";s:7:"classes";s:0:"";s:9:"thumbnail";s:0:"";s:9:"collapsed";s:0:"";s:9:"optimized";s:0:"";}
- kc_raw_content:
- [kc_row use_container="yes" _id="99244"][kc_column width="12/12" video_mute="no" _id="519644"][kc_spacing height="20" _id="797039"][kc_column_text _id="588872"]
If it is possible to build nanostructures of controlled shape by exploiting the double helix structure of DNA molecules (leading to assemblies commonly called "DNA origami"), it remains much more difficult to develop such structures. precise from proteins. However, in living cells, very sophisticated supramolecular architectures such as microtubules, actin filaments, or flagella perform vital functions and are entirely made up of natural proteins. The latter assemble spontaneously because each protein has a particular shape that allows it to interact in a very specific way with other proteins, thus leading to complex architectures. Creating ordered supramolecular architectures of proteins would open the door to many applications in biology but also in materials science.
[/kc_column_text][kc_spacing height="20" _id="437938"][/kc_column][/kc_row][kc_row use_container="yes" _id="465016"][kc_column width="40%" _id="891298"][kc_spacing height="20" _id="868830"][kc_single_image image_size="full" _id="444597" image_source="media_library" image="42384"][kc_spacing height="6px" _id="121493"][kc_column_text _id="586902"]Figure: Semi-experimental model of the origami of artificial proteins whose superhelix of bricks (in blue) self-assembles by affinity with the staple (in yellow). The extreme regularity of the supramolecular structure obtained arranges the staples periodically along the superhelix and induces the formation of aligned origami crystals (see background).
[/kc_column_text][kc_spacing height="10px" _id="948133"][kc_column_text _id="850874"]
@ I2BC, CEMES, CBI, IPR and ICB- Read the article of the INP
- Design, synthesis, and characterization of protein origami based on self-assembly of a brick and staple artificial protein pair. L. Moreaud et al., PNAS, published on March 09, 2023
DOI : 10.1073/pnas.2218428120
Four French teams demonstrate the possibility of building ordered supramolecular architectures that form spontaneously from proteins specially designed for this purpose. The key to this innovation is the engineering of highly regular proteins with repeating patterns, which have recognition surfaces allowing them to establish specific interactions.
[/kc_column_text][kc_column_text _id="751196"]The Laboratoire Interdisciplinaire Carnot de Bourgogne (ICB), the Centre d'élaboration de matériaux et d'études structurales (CEMES, CNRS), Institut de biologie intégrative de la cellule ( I2BC, CNRS / CEA / Université Paris Saclay), l'Institut de physique de Rennes (IPR, CNRS / Université de Rennes), and the Centre de biologie intégrative (CNRS / Université de Toulouse - Paul Sabatier) have developed a generalizable method for constructing artificial protein architectures. Rather than modifying natural proteins, scientists have favored the design of new proteins that are both very regular and programmed to assemble in precise geometries capable of forming stable superstructures. One of these proteins, called "staple", has the role of arranging precisely several other proteins called "bricks", their assembly naturally giving the three-dimensional supramolecular architecture its structural complexity. A first challenge was therefore to design these proteins and produce them. Then, by combining several techniques including X-ray scattering and electron cryomicroscopy, the researchers showed that the proteins assemble in a few minutes, at room temperature and according to the expected architecture. This constitutes a first experimental whose principle can be generalized to other molecular systems.
[/kc_column_text][kc_column_text _id="685074"]These results are published in the journal Proceedings of the National Academy of Science.
[/kc_column_text][/kc_column][/kc_row]
